Prof. Dr. Jens Boch, Dingbo Zhang
Prof. Dr. Jens Boch, Dingbo Zhang
Leibniz Universität Hannover, Institute of Plant Genetics, Dep. of Plant Biotechnology
Leibniz Universität Hannover, Institute of Plant Genetics, Dep. of Plant Biotechnology
Genome editing via CRISPR/Cas is an exciting technology for modern breeding of crop plants. The technology is much more precise than any other breeding method and opens novel opportunities to finally improve the safety of food plants. Our aim is to remove allergens from peanut and mustard which can be highly dangerous for allergic people.
In peanut (Arachis hypogaea), the 7S globulin Ara h 1 is an allergenic seed storage protein. Because peanut is an allotetraploid plant, four gene copies exist in its genome. Similarly, in mustard (Brassica juncea), the 2S albumin Bra j 1 is also an allergenic seed storage protein with four gene copies. To remove the allergenic potential, all gene copies have to be modified.
Genome Editing in peanut and mustard
The genome editing field is raidly evolving with multiple new technologies becoming available every year. We use a flexible and modular cloning system (MoClo) to easily adopt novel tools into our reperoire. The classical CRISPR/Cas9 system is used to completely delete the coding regions of ara h 1 and bra j 1 from peanut and mustard, respectively, or to trigger short frame shift mutations that abort protein production. To delete the genes, four different sgRNAs flanking each gene are used simultaneously. In contrast, only one sgRNA is required for point mutations, which makes this approach easier and faster to generate plants for further studies. A complete deletion will eventually produce safer plants, because all allergenic determinants are removed. As an alternative approach, base editing can precisely change individual DNA bases to alter one or several amino acids, e.g., within the allergenic epitopes identified by project 1.1, to lower the allergenic potential of the peanut and mustard proteins.
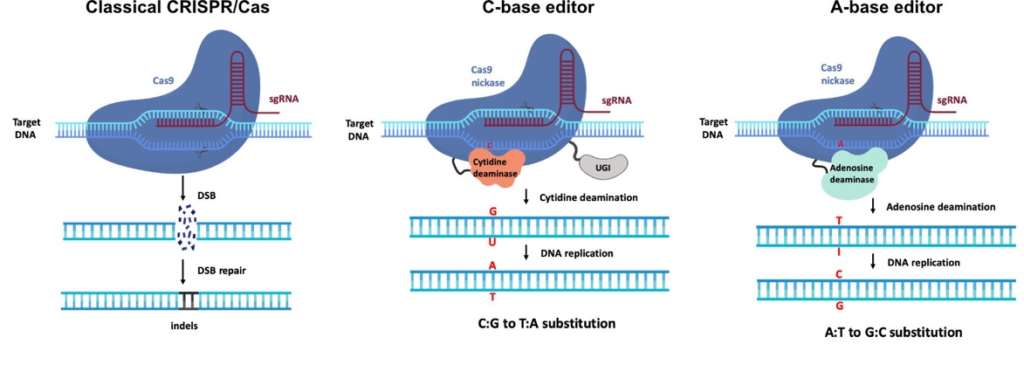
So far, we have established all tools, tested several sgRNA candidates, and chosen the most active ones for genome editing in peanut and mustard. We were able to generate target deletions in peanut protoplasts and frame shift mutations in mustard plants. Currently, we are establishing and analyzing editing events in whole plants in cooperation with subproject 2.